Contributions to Science
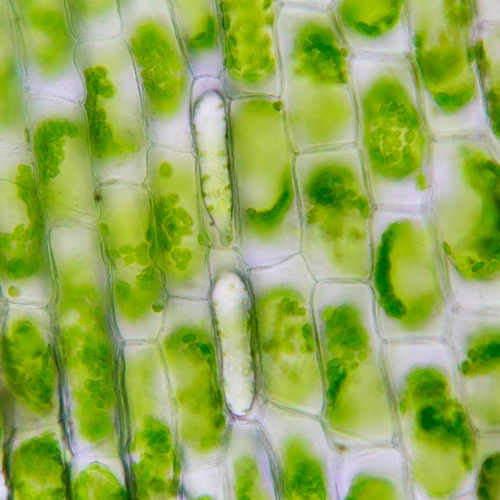
My early work in graduate school focused on understanding how plant cells separate, which is very rare because plant cells are connected by cell walls. However, cell separation occurs during male gametogenesis in many plants to create individual pollen grains. Through molecular genetics, cell biological, and biochemical approaches, I identified a class of mutants called quartet, which are required for cell separation, and subsequently determined the molecular nature of the defects through gene cloning and molecular and biochemical characterization. Using immunolocalization and biochemical analyses, I showed that the phenotpye resulted from defects in degrading the temporary cell wall before the secondary cell wall is deposited from the maternal tissue. I then cloned one of the genes, which encoded a pectin methylesterase, the first cell wall degrading enzyme with a demonstrated function in vivo. The quartet strains are still the de facto lines for plant scientists to study a variety of topics including gametophytic function, meiotic drive, genome stability, and centromere mapping. The strains have been used to map Arabidopsis centromeres, which was instrumental in refining the physical map and completing the genome sequencing. In the future, these strains could enable the creation of artificial plant chromosomes.
One of the biggest problems facing biology in the post-genome era is that we still do not know the functions of many genes (25%-75% of protein-encoding genes are not even predictable for their function based on sequence similarity), even for intensively studied organisms such as E. coli, yeast, and human. To systematically infer functions of genes and group them into pathways, my group collaborated with Dr. Ed Marcotte’s group to create the first plant genome-wide co-function network called AraNet. It can be used to systematically identify new genes in pathways and infer functions of uncharacterized genes based on the functions of their network neighbors. In addition to contributing to the design and analysis of the network, my group demonstrated that AraNet could be used successfully to guide the functional identification of novel genes. Using molecular genetic approaches, we discovered novel regulators of drought resistance and lateral root development, traits that are essential in engineering drought resistance in plants.
Membrane proteins are perhaps the darkest matter in the pool of uncharacterized proteins because of the difficulty of working with them biochemically and expressing them heterologously. To better understand how proteins function across and within membranes, my group collaborated with Dr. Wolf Frommer’s group to develop high-throughput experimental and computational pipelines to systematically identify interactions between membrane proteins and signaling proteins, testing over 6 million binary interactions between 3000 proteins. To date, this is still the largest eukaryotic membrane protein interaction network (such a network previously existed only for yeast, at ~10% of the scale). I led the bioinformatics component of the project where we created a computational pipeline to enable the large-scale experimental pipeline (primer design, sequence validation, and image and statistical analyses of the interactions) and analyzed the resulting protein interaction network. This is Last Updated September 9, 2021 4 a foundational resource for generating many new hypotheses. The vast majority of the membrane protein interactions we found had never before been identified. In addition, the methods we developed for generating high-throughput membrane protein interactions are applicable to any species and the datasets will be useful in identifying patterns of signaling and regulation in plants.
Transcriptional regulation is a fundamental process in biology and has been the subject of an intensive study. However, molecular, genetic, and evolutionary studies suggest that there must be additional layers of control that have not been discovered. To investigate into one of such layers, we used an integrated approach (applying concepts, data, and tools from computer science, genetics, genomics, proteomics, molecular evolution, development, and stress physiology) to uncover a new layer of transcriptional regulation across many domains of life. There are a handful of anecdotal examples of transcription factor-like proteins without a DNA binding Last Updated September 9, 2021 5 domain, coined microProteins (miPs), which regulate evolutionarily related transcription factors. To test the prevalence of this mechanism, my group developed a genome-scale platform to discover, classify, and validate microProteins in Arabidopsis. We found over 400 putative miPs in Arabidopsis along with their putative target transcription factors and their respective biological pathways. In collaboration with experimental biologists at Carnegie and Stanford, we experimentally validated two novel miPs and their predicted target transcription factors using genetic, molecular, and biochemical experiments as a proof-of-concept. Given the prevalence of miPs in Arabidopsis, we applied the same strategy to predict miPs from 19 species, ranging from microbes to plants and metazoans. We detected putative miPs in all organisms examined and paired them with potential targets in almost all known transcription factor families. Our analysis suggests a potential ubiquitous layer of transcriptional regulation by miPs and provides a systematic framework for their future study. The potential universality of miP function may offer new tools to modulate transcription factor function in practical applications ranging from gene therapy to bioengineering.
As genome sequencing became feasible towards the end of my graduate work, I became interested in the possibility of genome-enabled biology to understand the functions of all genes and pathways encoded in a genome and elucidate how organisms are hard- and soft-wired. As an early career investigator at Carnegie, I led a team of biologists and software engineers to create a computational infrastructure called the Arabidopsis Information Resource (TAIR) to collect and encode all available genomic and literature data to be computable by algorithms and easily accessible by researchers. TAIR has been a primer for revolutionizing plant research by enabling systematic and quantitative analyses of biological functions and pathways. Some 20,000 scientists around the world are still actively using it. In addition, my group was one of the early developers of the Gene Ontology (GO) system where we contributed to making the system to work for plant genomes. GO is a shared, controlled and structured vocabulary for describing gene attributes. GO has been instrumental in analyzing and interpreting genomic and post-genomic data across many organisms and has been used to analyze data in thousands of research articles, including many studies of various human diseases.
Plant metabolism plays a vital role in the health and well-being of our society. Despite our dependence on plants for energy, nutrition, and medicine, plant metabolism remains a surprisingly understudied field. For example, more than 30% of all pharmaceuticals are based on plant natural products, yet our knowledge of plant metabolic pathways accounts for less than 0.1% of the metabolites thought to exist in flowering plants. Understanding how plants evolved this prodigious chemical vocabulary has been a longstanding goal in plant biology. My group developed computational pipelines that systematically annotate enzyme function on the genome-scale. Using this system, we created a unique, unified resource of plant metabolic networks and discovered several properties that illustrate the differential evolution of secondary metabolism, permitting elucidation of novel secondary metabolic pathways. This opportunity is particularly relevant because secondary metabolites often confer upon plants the ability to survive major biotic and abiotic threats, and are the major sources of medicine, fragrance, and flavor. Thus, the molecular components involved in the production of secondary metabolites are a source of great interest across many fields of research, including agricultural biotechnology, synthetic biology, and biomedical and pharmaceutical research.
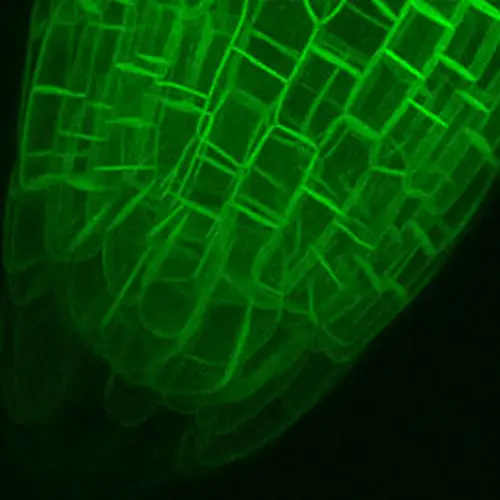
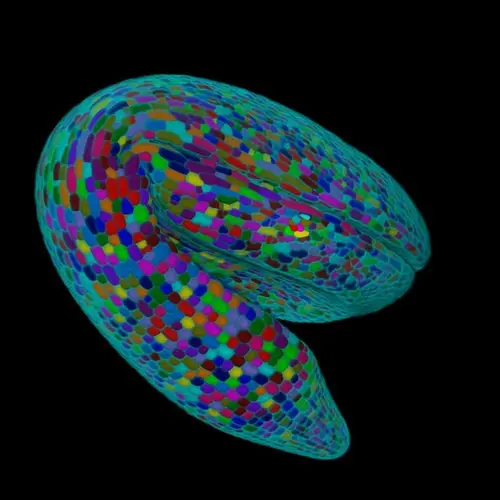
To supercharge our ability to understand how plants work, we need to quantitatively understand the dynamic molecular organization of plant cells and their functions. For this, we need a solid infrastructure that can incorporate and codify the theoretical and empirical data of plant cells, a task too big to tackle for a single group. Therefore, we want to create a community that includes scientists from plant biology, data science, AI, imaging, proteomics, single cell profiling and nanotechnology to lay the groundwork for creating a comprehensive understanding of the dynamic molecular organization of the plant cell, an initiative we are calling the Plant Cell Atlas (PCA). We have successfully kickstarted the PCA community building activities this spring with three digital workshops on the vision, technologies and broader impacts of the PCA. Because of COVID-19, our original plan for an in person gathering of 70 scientists, mostly senior faculty, turned into three virtual workshops, each of which drew over 400 scientists (70% early career) participating from around the world.
CV
- Ph.D., 1998: Stanford University, Molecular Genetic Analysis of Cell Separation during Arabidopsis thaliana pollen development
- B.A., 1992: Swarthmore College, Biology
- American Society of Plant Biologists (2010-present)
- International Society of Biocuration (2010-present)
- American Chemical Society (2014-present)
- Society for the Study of Evolution (2014-present)
- Society of Molecular Biology and Evolution (2014-present)
- Genetics Society of America (2014-present)
- International Society for Computational Biology (2015-present)
- California Native Plant Society (2015-present)
- Northern California Science Writers Association (2016-present)
- American Society of Cell Biologists (2016-present)
- American Geophysical Union (2019-present)
- NSF Predoctoral Fellowship (1993-1996)
- NSF/DOE/USDA Plant Training Grant Fellowship (1992-1993)
- Sigma Xi National Society (1991-1992)
- Howard Hughes Undergraduate Research Fellowship (1990-1991)
- National Honors Society (1988)
Scientific Advisory Boards:
DOE Biological and Environmental Research Advisory Committee’s Subcommittee Working Group on Biodesign
(2021-2022); Committee of Visitors, DOE Biological Systems Science Division (2021); Steering Committee, Plant
Cell Atlas (2021-2026); Advisory Committee, Joint Genome Institute (2020-present); Advisory Committee, Gene
Ontology Consortium (2019-present); Scientific Advisory Board, Phylos, Inc. (2018-present); ASPB Award
Nominations Committee (2018-present); Advisory Committee, IMPB conference (2018); Scientific Advisory
Board, VIB Department of Plant Systems Biology, Belgium (2016-present); Scientific Advisory Committee, Joint
Genome Institute’s Plant Group (2015-present); Scientific Advisor Board, Protein Data Bank (2009-present);
Advisor, Program for International Consortia and Collaboration on Agribioinformatics in National Agricultural
Genome Program (PICCAN) in Korea (2016-2017); Advisor, NSF C3-C4 Photosynthesis Project (2012-2013);
Member, Nominating Committee for the International Society of Biocuration’s Executive Committee (2009-2010);
Member, Nominating Committee for Plant Cyberinfrastructure Board of Directors (2007); Scientific Advisory
Committee, Value-directed Evolutionary Genomics Initiative (VEGI) (2010-2014); Scientific Advisory Committee,
CropLink Global Database (2006-2009); Steering Committee Member, International Solanaceae Genome
Initiative (2004-2008); Scientific Advisory Board, Saccharomyces Genome Database (SGD) (2003-2006);
Scientific Advisory Board, GrainGenes (2003-2006); Scientific Advisory Board, Cornell Genomics (2002-2006);
Scientific Advisory Committee, ChromDB (2001-2004)
Grant Review Boards:
DOE (2018); NSF (2021, 2020, 2018, 2016, 2015, 2014, 2012, 2011, 2008, 2006); USDA-ARS (2002); NHGRI
(2002)
International Conference Organization Boards:
Lead organizer, First Plant Cell Atlas Symposium (2021); Co-organizer, JGI Plant Secondary Metabolite
Workshop (2021); Scientific Organizing Committee, VIB conference Plant Science for Climate Emergency (2021); Lead organizer, First Plant Cell Atlas Workshop (2020); Co-organizer, 2nd Plant Systems Biology Conference (2020); Co-organizer, Plant Genomes, Systems Biology, and Engineering Conference at Cold Spring Harbor Laboratory (2021, 2019, 2017); Co-organizer, Forth Conference of International Society for Biocuration (2010); Lead organizer, Second International Biocurators meeting (2007); Co-organizer, Solanaceae Genomics meeting (2007); Lead organizer, First International Biocurators Conference (2005); Co-organizer, NSF sponsored workshop on ‘National Plant Synthesis Center’ (2005)
Scientific Journal Editorial Boards:
Advisory Editor, Plant & Cell Physiology (2020-2023); In silico Plants Editorial Board (2018-present); Associate Editor, Molecular Plant (2014-2019); Monitoring Editor, Plant Physiology (2002-2008, 2013-2016, 2021)
Press Releases
Recent Talks

Thriving In Extremes: A Virtual Conversation with Sue Rhee hosted by President Eric Isaacs
Sue Rhee, Plant Cell Atlas